Apr 16, 2024
Discovery of a group of plant RNA-induced silencing complexes that bind strongly to DNA
— A promising avenue for regulating gene expression —
Keyword:RESEARCH
OBJECTIVE.
Associate Professor Hiro-oki Iwakawa of the Rikkyo University Faculty of Science has discovered a group of plant RNA-induced silencing complexes (RISCs)*1 capable of binding strongly to DNA. His findings were published in the journal “Nucleic Acids Research” on April 16, 2024 (Japan time).
Key points
- Identification of differing nucleic acid binding properties among 3 groups of plant RNA-induced silencing complexes (RISCs)
- In one of the groups, RISCs that work in the nucleus appeared to bind strongly to DNA in addition to RNA, a previously unknown property
- Findings could aid in developing novel techniques for regulating gene expression
Study overview
In his research, Rikkyo University Associate Professor Hiro-oki Iwakawa revealed that the 3 groups of RISCs found in plants each have different nucleic acid binding properties. He showed that the 2 groups that function in the cytoplasm bind strongly to RNA, while the group that acts in the nucleus binds to DNA with equal or greater strength than it does to RNA. These findings suggest the existence of a previously unknown mechanism—that of plant RISCs directly binding to DNA—and could be of great help in developing new techniques for regulating gene expression.
This research was conducted using a grant from the Japan Science and Technology Agency's FOREST program (Fusion Oriented REsearch for disruptive Science and Technology) "Understanding and Application of Plant RNAi: Toward Flexible Artificial Genome Expression" (Principal Investigator: Hiro-oki Iwakawa).
Study description
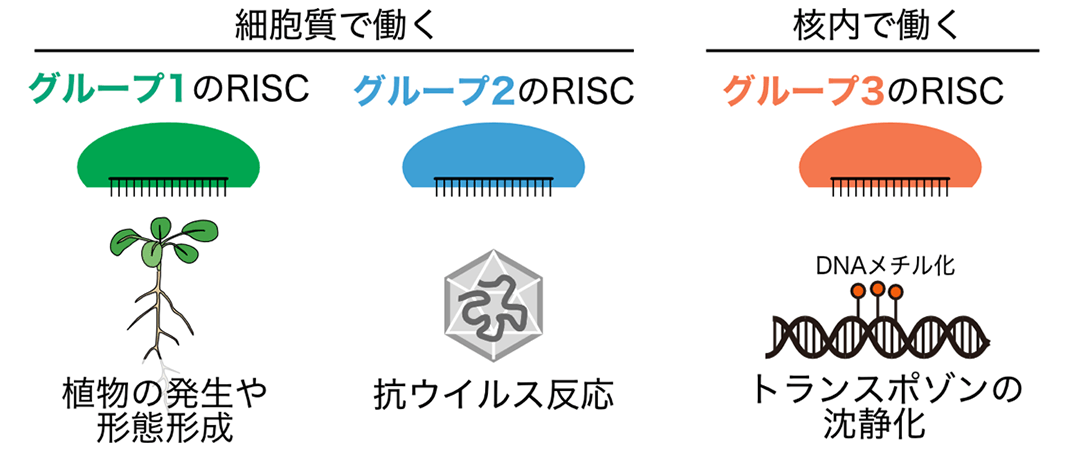
Figure 1: Typical functions of the RISCs in each group. Those in group 1 (especially AGO1-RISC) are important for plant development and morphogenesis, AGO2-RISC in group 2 is involved in antiviral responses, and the RISCs in group 3 are known to be involved in silencing transposons and non-self genes.
In his research, Rikkyo University Associate Professor Hiro-oki Iwakawa described the nucleic acid binding properties of 3 groups of RISCs in Arabidopsis thaliana. He started by synthesizing AGO proteins using a plant cell-free translation system*4, to which small RNA was added to form AGO1-RISC from group 1, AGO2-RISC from group 2, and AGO4-RISC, AGO6-RISC, and AGO9-RISC from group 3. Purified RISC was mixed with single-stranded RNA or DNA that is either completely complementary (base-paired) or partially mismatched (not base-paired) with the small RNA, and their binding affinity was quantified using a biochemical technique called filter binding assay*5 (Figure 2). This showed that compared to groups 1 and 2, group 3 RISC was capable of binding (tolerating mismatches) even in the presence of low complementarity of the 3' supplementary region*6 (Figure 3). Of even more interest, he found that group 1 and 2 RISCs, which function in the cytoplasm, bind strongly to RNA, while group 3 RISCs, which act in the nucleus, binds more strongly to DNA than to RNA (Figure 3). These findings suggest that the RISCs in each group have developed different target binding properties for executing their unique functions.
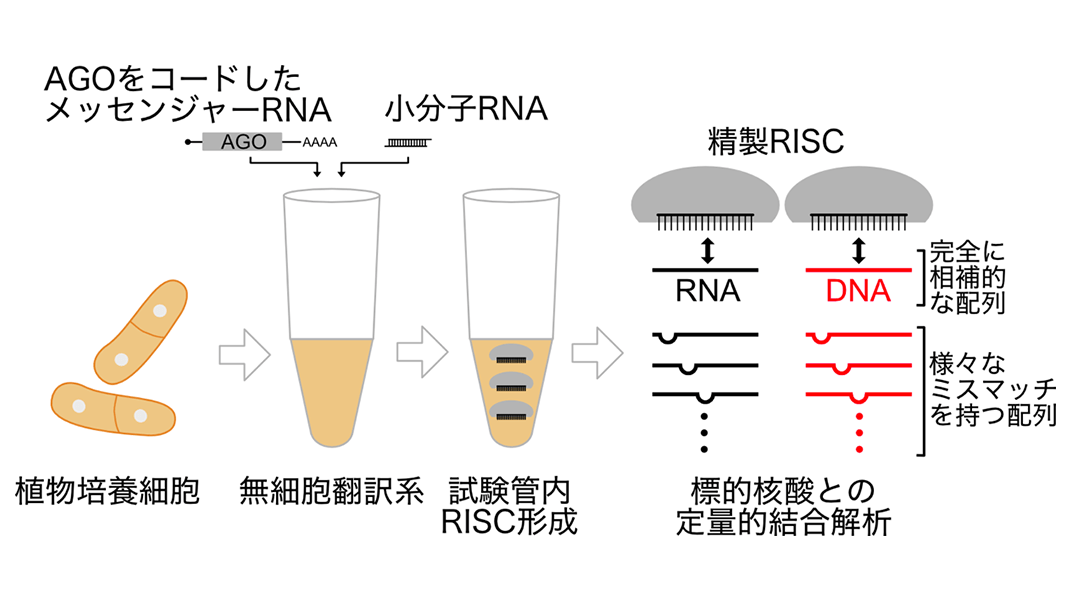
Figure 2: Experimental process. AGO proteins were synthesized in a cell-free translation system made from plant cultured cells, and the RISCs were formed by adding small RNA. Next, purified RISCs were mixed with nucleic acids (DNA/RNA) that possess sequences that were either completely complementary (base-paired) to the small RNA or had various mismatches. Quantitative binding analysis was performed using a method called filter binding assay.
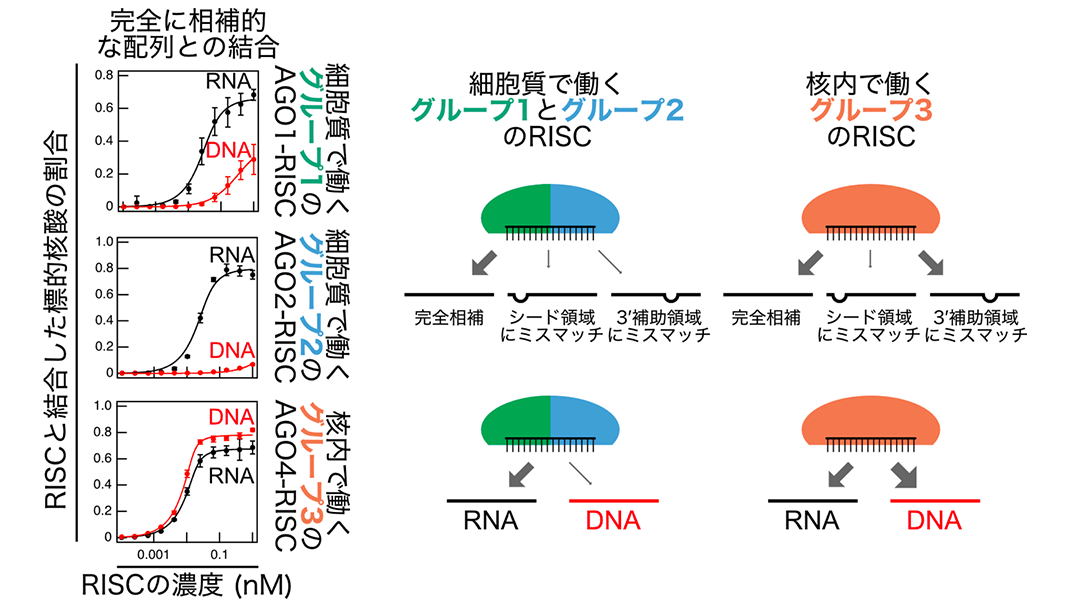
Figure 3: (Left) Examples of findings on quantifying binding of RISCs from each group to target RNA (black) and target DNA (red). (Top right) Compared to group 1 and 2 RISCs, which function in the cytoplasm, RISCs from group 3, which act in the nucleus, better tolerate mismatches in the 3' supplementary region. (Bottom right) The results also revealed that group 1 and 2 RISCs bind strongly to RNA, while group 3 RISCs bind to DNA as strongly as or more strongly than to RNA.
Publication information
Article title: The clade-specific target recognition mechanisms of plant RISCs
Author: Hiro-oki Iwakawa
DOI number: 10.1093/nar/gkae257
Research funding
Terminology
- *1 RNA-induced silencing complex (RISC):
A complex consisting of a single-strand small RNA and an argonaute (AGO) protein. When the small RNA part of the complex binds to complementary RNA, it specifically suppresses the expression of target genes.
- *2 Plasmids:
A general term for self-replicating DNA molecules that exist within bacteria and yeast cells, which differ from chromosomal DNA.
- *3 Phages:
A general term for viruses that infect and replicate inside of bacteria and archaea.
- *4 Plant cell-free translation system:
An experimental system in which a translation reaction is carried out in a test tube by adding amino acids, ATP, and other substances to an extract of cultured plant cells. By adding fabricated messenger RNA, it is possible to synthesize any protein a researcher wants in a test tube. Because a cell extract is used, biochemical reactions besides translation reactions can also be reproduced. In the present study, a novel tobacco cell-free translation system was used to synthesize AGO proteins and form the RISCs.
- *5 Filter binding assay:
A method for analyzing the binding affinity of two molecules that uses a membrane filter. The present study analyzed the binding affinity between RISCs and radioactively labeled nucleic acids (RNA or DNA).
- *6 3′ supplementary region:
The small RNA that exists in RISCs can be divided into 4 regions: the seed region (2 to 7-8 nucleotides from the 5′ end of the small RNA), central region (9-12 nucleotides), = 3′ supplementary region (13-16 nucleotides), and tail region (17th and on nucleotides). Previous research on animal RISCs have found that base pairings between the seed region and target RNA is essential for RISCs to bind to their targets, and that base pairings between the 3′ supplementary region and target RNA strengthens bonds between RISCs and their targets.