Apr 28, 2021PRESS RELEASE
Splitting the Escherichia coli genome into three parts to create new cells
- Genome installation technology for model organisms -
Keyword:RESEARCH
OBJECTIVE.
A research group led by Assistant Prof. Takahito Mukai and Prof. Masayuki Su’etsugu of the College of Science, Rikkyo University, has developed a technique for splitting and installing Escherichia coli genomes. Their research was conducted as part of the JST Strategic Basic Research Programs.
The E. coli genome consists of a single circular DNA with 4.6 million base pairs. Conventionally, this has been too large to be extracted from an E. coli cell for manipulation and transplantation into another E. coli cell. In their study, the group succeeded in reducing the size of the E. coli genome by splitting it into three parts (each with 1 million base pairs) and developed a technique for extracting the split genome from an E. coli cell and installing it in another cell. Their findings could be used not only to better understand the mechanism of genome replication and partitioning, but also as a synthetic biology tool for replacing genomes—the blueprints of life—to create functionally designed organisms.
The results of their research were published online as a Breakthrough Article in the British scientific journal "Nucleic Acids Research" on April 28, 2021 (UK time).
Main points
- The researchers maintained an E. coli genome as 3 pieces of circular DNA consisting of 1 million base pairs each.
- They developed a technique for extracting the split genome from an E. coli cell and transplanting it into another cell.
- Future applications of this technique are anticipated in synthetic biology, such as for transplanting an artificially synthesized split genome or constructing artificial E. coli designed to have useful functions.
JST Strategic Basic Research Programs team research (Core Research for Evolutional Science and Technology: CREST)
- Research field: "Creation of cell control technology using genome-scale DNA design and synthesis" (Research supervisor: Prof. Haruhiko Shiomi, Keio University School of Medicine)
- Research topic: "Cell-free on-chip synthesis and activation of artificial genomes"
- Principal Investigator: Masayuki Su’etsugu (Professor, Rikkyo University College of Science)
- Research period: October 2018 - March 2023
In this study, the group developed a cell-free on-chip method of genome synthesis that uses in vitro reconstruction techniques and microfabrication/manipulation techniques. They evaluated the process of transplanting a synthetic genome into cells and booting-up it, with the objective of developing a high-speed, low-cost cycle of "genome-synthesis, booting-up, evaluation." Using the genome synthesis techniques they developed, they plan to proceed with minimizing and defragmenting the E. coli genome to construct a "minimum platform cell" that simplifies the genome to a level that is completely understandable.
Study background and process
However, the organisms with genomes that can be installed with this method are still limited to mycoplasma, and there have been no successes with model organisms that are widely used in research and industry, such as E. coli. One challenge is the genome size. The E. coli genome, at 4.6 million base pairs, is more than four times larger than the mycoplasma genome, and its physical length is 2 millimeters, which is nearly 1,000 times the size of an E. coli cell. Although it is compactly folded inside the cell, it becomes severed and broken as soon as it is taken outside.
This group investigated ways of splitting the E. coli genome and maintaining it as multiple small chromosomes as a means of achieving genome installation. In E. coli and many other bacteria, the genome is encoded by a single large circular chromosome, though in eukaryotes this is encoded and divided into multiple chromosomes. If the chromosomes could be divided into pieces similar to mycoplasma size, the split chromosomes could be taken in and out of E. coli cells and installed in other cells to create artificial E. coli. This is what the group envisioned as they set out on their research.
Content of the study
Next, they investigated a method for extracting and purifying the three split chromosomes with 1 million base pairs each from the E. coli as supercoiled DNA2. Surprisingly, the purified split chromosome could be introduced into a different E. coli strain with natural chromosomes consisting of 4.6 million base pairs using electroporation, a classical method of creating pores in a cell membrane using electrical pulses to introduce DNA (Figure 1). This is significantly larger than the DNA that had been previously introduced into an E. coli cell. In addition, they showed it is possible to exchange split chromosomes between E. coli strains that have three split chromosomes.
Future prospects
In this study, researchers for the first time succeeded in extracting genome-sized DNA with 1 million base pairs from E. coli and installing it in another cell. The group is also developing a technique for synthesizing large DNA outside of cells (cell-free) and has reported on a technique for cell-free amplification of circular DNA with 1 million base pairs. In the future, the ability to install split chromosomes synthesized using cell-free methods into E. coli cells could help create artificial E. coli designed to have useful functions, such as the ability to produce certain substances.
Figure
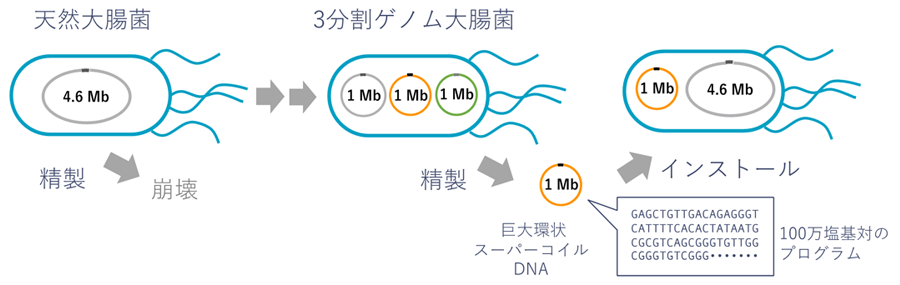
Figure 1. Creation of a three-part Escherichia coli genome and installation of the split genome
Glossary
The ori-par system consists of a DNA region (ori) that encodes the replication initiation sequence and a DNA region (par) that partitions the DNA after replication. Together they stably maintain large DNA in bacteria that proliferate via cell division. The sop partitioning system is a par system derived from the E. coli F plasmid.
2) Circular supercoil DNA
A form of circular double-stranded DNA (closed-ring DNA) twisted into a compact state.