Mar 16, 2020PRESS RELEASE
Comparative genomics exploring the origins of land plants, successful high-precision sequencing of hornwort genomes
Keyword:RESEARCH
OBJECTIVE.
Rikkyo University, Kanazawa University, and Hiroshima University are pleased to announce the successful sequencing of multiple hornwort genomes, filling in a missing piece of comparative genomics in the search for the origins of land plants.
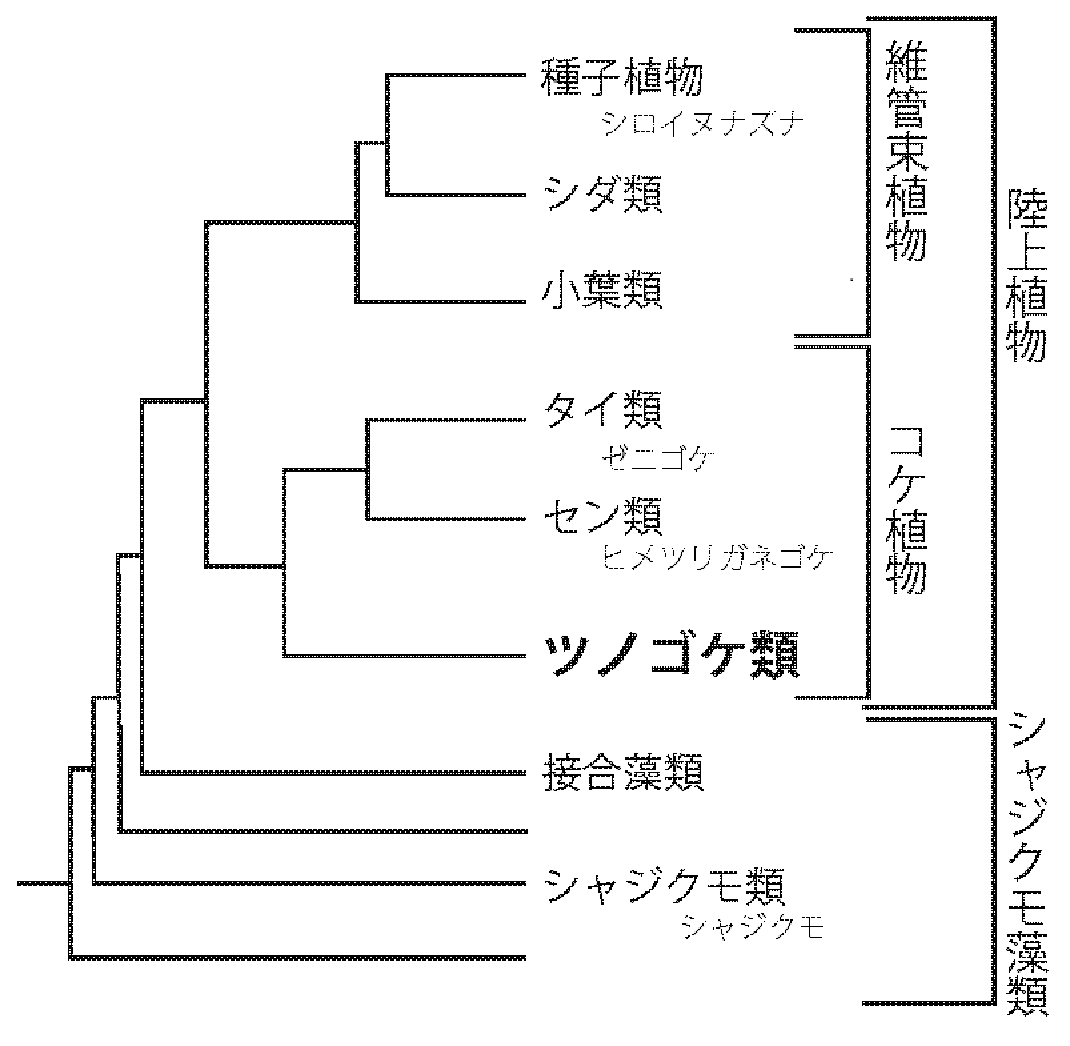
Figure 1: Phylogenetic relationships of land plants
Associate Professor Keiko Sakakibara of the Department of Life Science at Rikkyo University, Assistant Professor Tomoaki Nishiyama of the Advanced Science Research Center at Kanazawa University, and Associate Professor Masaki Shimamura of the Graduate School of Integrated Sciences for Life at Hiroshima University were part of a joint international research group that decoded three genomes from two hornwort species. Other members of the team included Assistant Professor Fay-Wei Li of the Boyce Thompson Institute in the United States (Li is also assistant professor at Cornell University) and Peter Szovenyi, lecturer at the Zurich-Basel Plant Science Center in Switzerland. Hornworts split off around the time that mosses and liverworts were diverging from vascular plants, and are thus a key lineage for thinking about the earliest stages of land plant evolution. Genomes for members of these other categories have already been published. Molecular phylogenetic analysis of the results of this study demonstrates that hornworts along with mosses + liverworts (monophyletic group comprising mosses and liverworts) form a monophyletic group that includes all bryophytes (Figure 1). This genome sequencing also advances the understanding of the genetic basis for some of the unique characteristics of hornworts.
The study's results were published in the scientific journal "Nature Plants" (1 a.m. Japan time on March 14, 2020, 4 p.m. U.K. time on March 13) (title: Anthoceros genomes illuminate the origin of land plants and the unique biology of hornworts).
The study's results were published in the scientific journal "Nature Plants" (1 a.m. Japan time on March 14, 2020, 4 p.m. U.K. time on March 13) (title: Anthoceros genomes illuminate the origin of land plants and the unique biology of hornworts).
1. Paper background
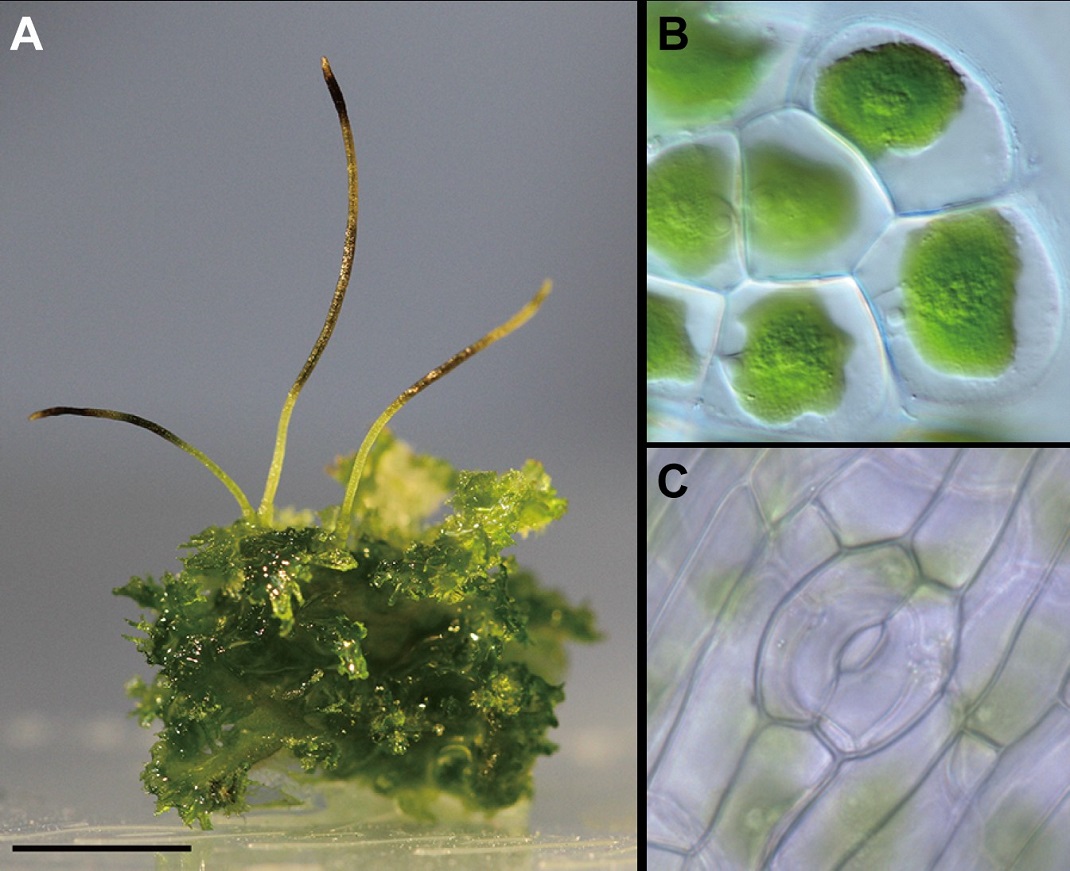
Figure 2: Images of the hornwort Anthoceros agrestis
Land plants are divided into vascular plants, which possess vascular bundles made up of tracheids or vessels with lignified*1 secondary cell wall thickening, and bryophytes, which form a single sporangium in the sporophyte without lignified secondary cell wall thickening. Hornworts are bryophytes along with mosses and liverworts. Hornworts have gametophytes with flat, leaf-like structures and nitrogen-fixing cyanobacteria that exist symbiotically in internal cavities. They also exhibit a unique horn-shaped sporophyte morphology with a meristem at the base (Fig. 2A). In addition, similar to green algae, hornworts usually have only one or two chloroplasts per cell and have a pyrenoid-based*2 carbon concentrating mechanism (CCM), which is characteristic of green algae (Fig. 2B). Because hornworts have many exceptional properties even among bryophytes, there has been debate over whether they were the first to diverge from land plants, are closely related to vascular plants, or form a monophyletic group of bryophytes along with mosses and liverworts. The genome information of the moss Physcomitrium patens was published in 2008 and that of the liverwort Marchantia polymorpha in 2016. These illuminated the genetic composition of common ancestors of land plants in the early stages of evolution, but until this year there had been no reports on hornworts. The first genomes for this important group of land plants were published this year, that of Anthoceros angustus in February by a Chinese research group and the genomes revealed in the present study.
2. Results of the study
This study published the genomic sequences and annotations (analyses of which genes are where) of three strains of two hornwort species (Anthoceros agrestis Bonn and Oxford isolates, Anthoceros punctatus). The genome sequences contain from 117 to 133 million base pairs. For Anthoceros agrestis Bonn, the Hi-C method (a technique for estimating physical proximity between chromosomal regions in organisms) identified six large scaffolds (sections of DNA sequences that can be ordered even if some areas are unknown) and small scaffolds. This number is consistent with the results of chromosomal fluorescence studies, which shows that the base sequences were successfully reconstructed at the chromosomal scale. Generally, plant genomes have many repetitive sequences congregated near the kinetochore, but this pattern was not found in the chromosome-scale assemblies of hornworts. This was the second genome to be discovered with a pattern that lacks an abundance of these repetitive sequences near the kinetochore, after that of Physcomitrium patens. Orthologs*3 were estimated based on a dataset that included Streptophyte algae (Chara braunii and Zygnematales), the moss Physcomitrium patens, and the liverwort Marchantia polymorpha, with molecular phylogenetic analysis strongly indicating that hornworts form a monophyletic group with mosses and liverworts, and are a sister group. In other words, this provides more evidence that the bryophytes are a monophyletic group, which has been supported by recent molecular phylogenetic studies.
Unlike other bryophytes, hornworts have an persistent meristem at the base of the sporophyte, which some say is similar to the sporophytes of vascular plants that also have an indeterminate meristem. This is why there has long been support for the theory that hornworts are closely related to vascular plants. One of the genes that functions to maintain the sporophyte meristem in vascular plants is KNOX1. However, this study shows that the KNOX1 gene was not found in the hornwort genomes. Rather, there is the possibility that the meristem of the hornwort sporophyte is regulated by a different mechanism than in vascular plants, and forms in a unique manner.
In mosses, hornworts, and vascular plants, specialized cells called guard cells differentiate in the epidermis to form stomata leading to inside the plant (Fig. 2C). The genome sequencing in this study revealed that the gene repertoire that regulates guard cell differentiation in hornworts is also present like Physcomitrium patens (moss) and Arabidopsis thaliana (vascular plant). This shows the homology of stomata between vascular plants and hornworts at the genetic level. While the lack of stomata with guard cells in liverworts has been thought to indicate the early divergence from land plants, in fact liverworts lost the genes for stomata formation.
Hornworts are also known to exhibit symbiotic nitrogen-fixing cyanobacteria. Candidate genes involved in this symbiosis have been identified by genome sequencing and RNA-seq (cDNA sequence analysis using next-generation sequencers). This study compared gene expression in a hornwort cultured with and without the cyanobacterium Nostoc punctiforme. The genes that exhibited different expression levels included receptor kinases, transcription factors, and transporters. One that deserves attention is a SWEET sugar transporter in the SWEET16/17 clade. This is thought to enable the hornwort to secrete sugar that is supplied to cyanobacteria during symbiosis, which enables nitrogen fixation. SWEET sugar transporters are also used in symbiosis with mycorrhizal fungi, but this uses another gene belonging to the SWEET1 group, which indicates symbiosis with cyanobacteria and mycorrhizal fungi evolved independently.
The Earth's atmosphere currently has low levels of carbon dioxide. How plants collect CO2 is an important factor in photosynthesis. Green algae and some hornworts have a structure called a pyrenoid in their chloroplasts, which has a mechanism for concentrating carbon dioxide. The hornwort genomes were searched for homologous genes with the CCM gene group in the green algae Chlamydomonas reinhardtii. This only identified LCIB, which is not found in other land plants. The research group believes this gene is associated with the pyrenoid-based CCM in hornworts. The genomic information on hornworts published in this study is expected serve as the basis for further comparative studies with land plants and evolutionary genetic research on hornworts.
Tomoaki Nishiyama and Keiko Sakakibara drafted the research proposal, sequenced the genomes, and wrote the manuscript. Masaki Shimamura analyzed the chromosomes of the hornworts. Keiko Sakakibara analyzed the transcription factors. Tomoaki Nishiyama annotated the genomes and analyzed the genes related to stomata formation.
【Glossary】
*1) Lignin
A polymer phenolic compound found in the tracheids, vessels, and secondary cell walls of the vascular bundles of vascular plants.
*2) Pyrenoid
A structure in the chloroplasts of algae with RuBisCO crystals that catalyze carbon dioxide fixation. Many are surrounded by reserve substances such as starch.
*3) Ortholog
Genes from different organisms that were the same gene in a common ancestor.
Unlike other bryophytes, hornworts have an persistent meristem at the base of the sporophyte, which some say is similar to the sporophytes of vascular plants that also have an indeterminate meristem. This is why there has long been support for the theory that hornworts are closely related to vascular plants. One of the genes that functions to maintain the sporophyte meristem in vascular plants is KNOX1. However, this study shows that the KNOX1 gene was not found in the hornwort genomes. Rather, there is the possibility that the meristem of the hornwort sporophyte is regulated by a different mechanism than in vascular plants, and forms in a unique manner.
In mosses, hornworts, and vascular plants, specialized cells called guard cells differentiate in the epidermis to form stomata leading to inside the plant (Fig. 2C). The genome sequencing in this study revealed that the gene repertoire that regulates guard cell differentiation in hornworts is also present like Physcomitrium patens (moss) and Arabidopsis thaliana (vascular plant). This shows the homology of stomata between vascular plants and hornworts at the genetic level. While the lack of stomata with guard cells in liverworts has been thought to indicate the early divergence from land plants, in fact liverworts lost the genes for stomata formation.
Hornworts are also known to exhibit symbiotic nitrogen-fixing cyanobacteria. Candidate genes involved in this symbiosis have been identified by genome sequencing and RNA-seq (cDNA sequence analysis using next-generation sequencers). This study compared gene expression in a hornwort cultured with and without the cyanobacterium Nostoc punctiforme. The genes that exhibited different expression levels included receptor kinases, transcription factors, and transporters. One that deserves attention is a SWEET sugar transporter in the SWEET16/17 clade. This is thought to enable the hornwort to secrete sugar that is supplied to cyanobacteria during symbiosis, which enables nitrogen fixation. SWEET sugar transporters are also used in symbiosis with mycorrhizal fungi, but this uses another gene belonging to the SWEET1 group, which indicates symbiosis with cyanobacteria and mycorrhizal fungi evolved independently.
The Earth's atmosphere currently has low levels of carbon dioxide. How plants collect CO2 is an important factor in photosynthesis. Green algae and some hornworts have a structure called a pyrenoid in their chloroplasts, which has a mechanism for concentrating carbon dioxide. The hornwort genomes were searched for homologous genes with the CCM gene group in the green algae Chlamydomonas reinhardtii. This only identified LCIB, which is not found in other land plants. The research group believes this gene is associated with the pyrenoid-based CCM in hornworts. The genomic information on hornworts published in this study is expected serve as the basis for further comparative studies with land plants and evolutionary genetic research on hornworts.
Tomoaki Nishiyama and Keiko Sakakibara drafted the research proposal, sequenced the genomes, and wrote the manuscript. Masaki Shimamura analyzed the chromosomes of the hornworts. Keiko Sakakibara analyzed the transcription factors. Tomoaki Nishiyama annotated the genomes and analyzed the genes related to stomata formation.
【Glossary】
*1) Lignin
A polymer phenolic compound found in the tracheids, vessels, and secondary cell walls of the vascular bundles of vascular plants.
*2) Pyrenoid
A structure in the chloroplasts of algae with RuBisCO crystals that catalyze carbon dioxide fixation. Many are surrounded by reserve substances such as starch.
*3) Ortholog
Genes from different organisms that were the same gene in a common ancestor.
3. Published article
- Journal name: Nature Plants
- Title: "Anthoceros genomes illuminate the origin of land plants and the unique biology of hornworts"
- Authors: Li, F.-W., Nishiyama, T., Waller, M., Frangedakis, E., Keller, J., Li, Z., Fernandez-Pozo, N., Barker, M. S., Bennett, T., Blázquez, M. A., Cheng, S., Cuming, A. C., de Vries, J., de Vries, S., Delaux, P.-M., Diop, I. S., Harrison, J., Hauser, D., Hernández-García, J., Kirbis, A., Meeks, J. C., Monte, I., Mutte, S. K., Neubauer, A., Quandt, D., Robison, T., Shimamura, M., Rensing, S. A., Villarreal, J. C., Weijers, D., Wicke, S., Wong, G. K.-S., Sakakibara, K., Szövényi, P.
4. Other
- This project was supported by the National Institute for Basic Biology (NIBB) Collaborative Research Program grant no. 13-710 to Tomoaki Nishiyama; the Japan Society for the Promotion of Science (JSPS) grant no. KAKENHI 15H04413 to Tomoaki Nishiyama; the JSPS grant nos. KAKENHI 25113001, 26650143, 18H04843 and 18K06367 to Keiko Sakakibara, and by other research grants from various countries.