News
Rikkyo researchers propagate large circular DNA by reconstituting multiple cycle of genome replication in vitro
10/26/2017
A team of Rikkyo University researchers, led by Associate Professor Masayuki Suetsugu, has succeeded for the first time in reconstituting continuous repetition of the replication cycle of a bacterial genome. The research was conducted as part of a research and development project led by Hiroyuki Noji, a program manager of the Impulsing Paradigm Change through Disruptive Technologies Program (ImPACT), which is under the auspices of the Cabinet Office’s Council for Science, Technology and Innovation.
Research highlights:
1.Twenty-five types of protein are used to reconstitute Escherichia coli (E. coli) genome replication cycle in vitro for the first time.
2.E. coli genome replication cycle repeats autonomously and continuously in isothermal conditions, enabling amplification of large, circular DNA with more than 200,000 base pairs exponentially.
3.The developed technology enables cell-free DNA cloning easily without relying on E. coli.
Research outline
A team of Rikkyo University researchers, led by Associate Professor Masayuki Suetsugu, has succeeded for the first time in reconstituting continuous repetition of the replication cycle of bacterial genome. The research was conducted as part of a research and development project led by Hiroyuki Noji, a program manager of the Impulsing Paradigm Change through Disruptive Technologies Program (ImPACT), which is under the auspices of the Cabinet Office’s Council for Science, Technology and Innovation. Suetsugu is a former researcher for the Strategic Creative Research Promotion Project (PRESTO) of the Japan Science and Technology Agency (JST).
.
The replication cycle reconstitution system induces the exponential propagation of a single long-chain, circular DNA molecule with a replication origin. Conventionally, cloning larege DNA required E. coli cells as a host, but this new technology enables cell-free propagation of long-chain DNA with more than 200,000 base pairs (approximately 200 genes) only through isothermal reactions taking place over several hours. This development is important to elucidate the genome replication mechanism, and it also is expected to be a novel useful tool to artificially synthesize large DNA at the genome level.
.
An article detailing this research was published in the online edition of Nucleic Acids Research, a British science journal on September 28, 2017.
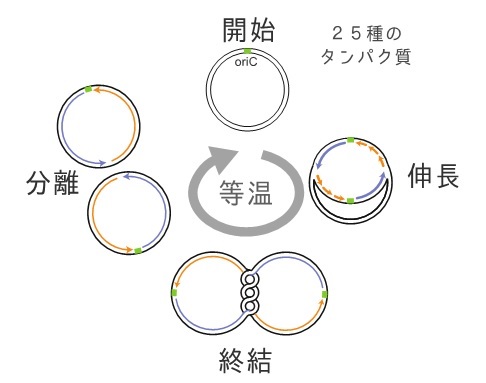
Figure 1: Replication Cycle Reaction
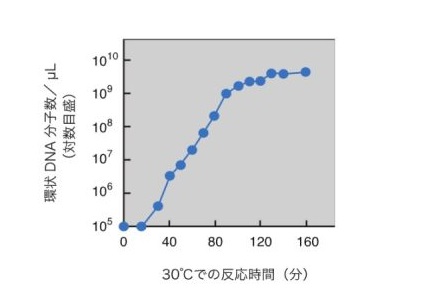
Figure 2: Exponential amplification of circular DNA molecules in replication cycle reaction
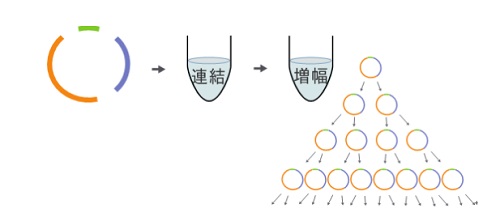
Figure 3: Cell-free cloning